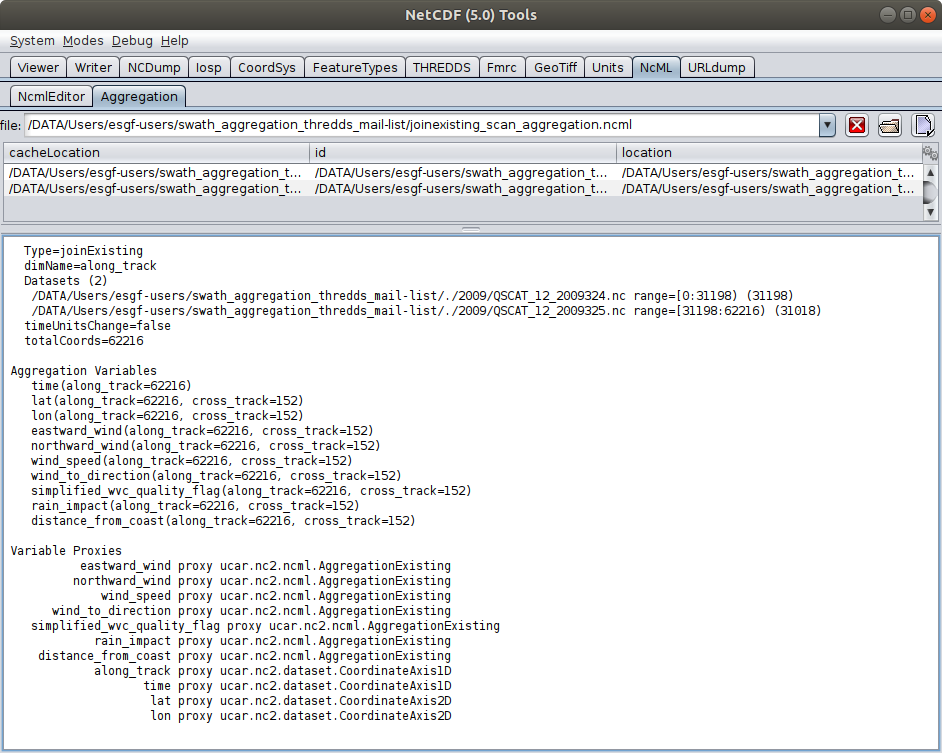
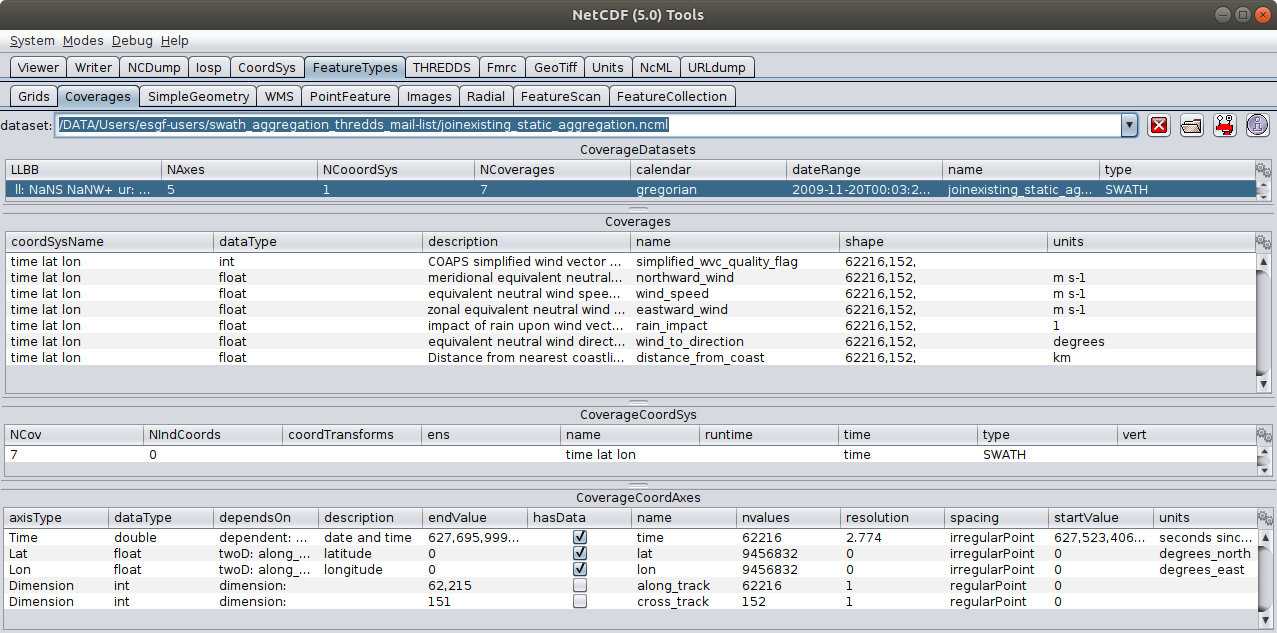
- To: Michael McDonald <mcdonald@xxxxxxxxxxxxx>
- Subject: Re: [thredds] ncml/THREDDS Aggregation of Swath Data follow up
- From: Antonio S. Cofiño <cofinoa@xxxxxxxxx>
- Date: Thu, 14 May 2020 13:50:57 +0200
- References:
- [thredds] ncml/THREDDS Aggregation of Swath Data follow up
- From: Ian Dimitri
- Re: [thredds] ncml/THREDDS Aggregation of Swath Data follow up
- From: Antonio S. Cofiño
- Re: [thredds] ncml/THREDDS Aggregation of Swath Data follow up
- From: Michael McDonald
- [thredds] ncml/THREDDS Aggregation of Swath Data follow up